Storage and processing of data
General recommendations for the storage of research data are:
- Naming convention: use short, meaningful file and folder names for easy identification.
- Folder structure: Organize the data logically in a clear, uniform folder structure.
- Data formats: Choose open, documented, standard formats recommended by the subject community to ensure long-term readability.
- Metadata: Capture sufficient standardized metadata to enable interpretability and reproducibility.
- Checksums: Calculate checksums (e.g. MD5 or SHA256) for the files to verify data integrity.
- Versioning: Use a versioning system (e.g. Git) to track changes to data and metadata.
- Storage location: Choose a long-term archiving system that ensures permanent data integrity and access.
Create checksum
1. press Windows+R to open the Run window.
2. enter cmd and click OK.
3. the Command Prompt window opens.
4. execute the following command:
certutil -hashfile C:\path\my_file.tif SHA256
Info: A path with spaces must be enclosed in quotation marks.
Format for storage and long-term archiving
Biological research generates a wide range of data from devices such as microscopes, sequencers and mass spectrometers. Each of these instruments often uses its own (proprietary) data formats, which can often only be read with special software. However, this makes data exchange, further processing of the data and long-term use or long-term archiving of the data extremely difficult.
In such cases, we recommend additional conversion of the data into a common data format (standard format). Standard formats are characterized by publicly accessible specifications, manufacturer independence and broad software compatibility.
Standard formats for microscopy data
Please use the most common data formats for storing and archiving microscopy data. Avoid the proprietary formats of the manufacturers, as well as JPEG or PNG. Use instead:
OME-TIFF: Recommended if OME-XML metadata is to be saved directly in the image file.
- Advantages: Standardized metadata structure (OME-XML), optimized for large image data sets, supports multidimensional data, widely used.
- Disadvantage: Not optimal S3 access.
OME-Zarr: Object-based format for large data sets and cloud-based workflows.
- Advantages: Combination of OME-XML metadata and efficient storage for fast access - whether local or online ("in the cloud"), scalability, S3-optimized (e.g. Amazon Simple Storage Service or comparable object storage), parallel access, support for large data sets and metadata.
- Disadvantage: Not ideal for very small data sets. Splitting into many small files can lead to problems on classic hard disks and file systems.
HDF5 (Hierarchical Data Format version 5): Suitable for very large data sets and multidimensional data.
- Advantages: Efficient storage of large amounts of data, supports metadata, flexible.
- Disadvantage: Not optimal S3 access.
Options for saving metadata
Metadata provides information about the creation, content and subsequent use of research data. It is often referred to as "data about data" and is crucial for the quality and integrity of research data. Although it represents independent data, it is often saved together with the actual research data.
There are different options for metadata storage:
- Embedded metadata: Storage of metadata directly in the data file (e.g. image).
- Sidecar metadata: Storage of metadata in separate files.
- Databases: Storage of metadata in a corresponding database (e.g. OMERO, openBIS) that is linked to the data.
Use of subject-specific metadata standards
Metadata includes technical and subject-specific data. Technical metadata (e.g. data volume, data format) plays a role in long-term data storage. Subject-specific metadata (e.g. technical terms, domain-specific knowledge or context information) ensures the comprehensibility and reuse of research data.
The specific requirements for metadata vary depending on the discipline and are defined in so-called subject-specific metadata standards, which can differ in terms of scope, vocabulary, structure and/or format.
Tips
- If possible, standard data and controlled vocabularies or ontologies should be used to describe the data.
- Early collection of metadata reduces the documentation effort at the end of the project.
- Repositories and journals may have specific requirements for data and metadata to be archived.
Important metadata standards in biological research
Image data:
- OME-XML (Open Microscopy Environment XML): Description of the technical metadata of an imaging experiment
- REMBI (Recommended Metadata for Biological Images)
- MITI (Minimum Information for Highly Multiplexed Tissue Imaging)
- MIHCSME (Minimum Information for High Content Screening Microscopy Experiments)
Sequence data:
- FASTA/FASTQ: Standard formats for DNA/RNA sequences and associated quality data.
- SAM/BAM/CRAM: For aligned sequence data.
- MIxS/MIMARKS: Minimum metadata standards for different types of sequence and metadata.
Bioinformatics and omics:
- MIAME (Minimum Information About Microarray Experiments)
- MINSEQE (Minimum Information about high throughput Sequencing Experiments)
- ISA-Tab / ISA-JSON: Framework for complex experimental studies (e.g. omics-based studies).
Please use the infrastructure provided by the IT of the School of Biology/Chemistry for storing and archiving digital research data:
Alternatively or additionally, please only use recognized national or international (subject-specific) repositories or archives.
The retention period for research data and records is at least ten years either after publication of the research data or from publication of the research results or after completion of the respective research activity. Deviations may result from legal or contractual regulations, from requirements of third-party funding bodies or internal guidelines.
Further open offers
GitLab - Repository for software code (Osnabrück University)
Chemotion - Electronic lab notebook for chemists (University of Braunschweig)
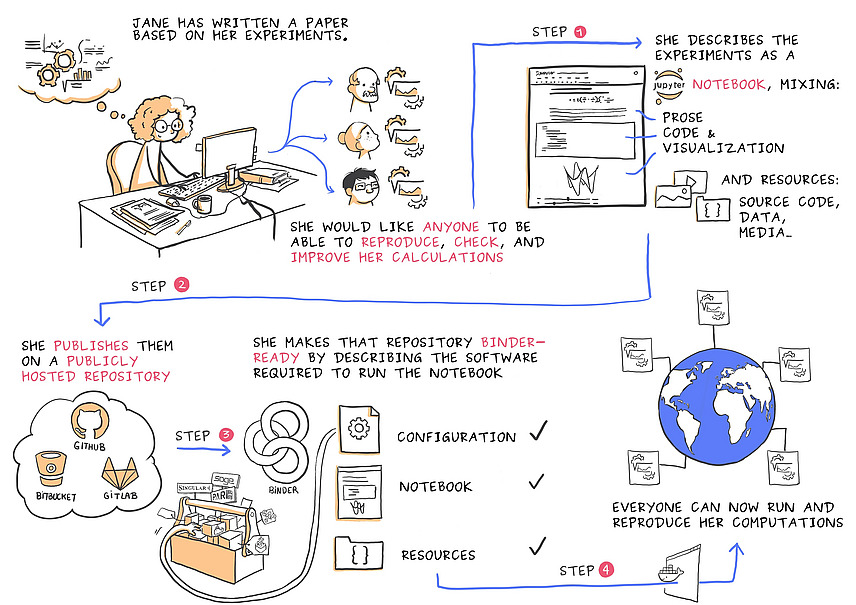
Software-supported processing and analysis of research data makes it possible to evaluate this data, but this process also requires comprehensive data management, including metadata, versioning and secure archiving, in order to ensure the reproducibility and usability of the research data.
Tips:
- For image data, use a direct link from Fiji/ImageJ and OMERO to automatically link your results.
- Use tools such as Jupyter Notebook or the Macrorecorder from Fiji which, in addition to automation, also ensure reproducible documentation of your analysis workflow, and link them to your data.